
Yonsei Biochem & CS
Sunghyun Park
Biography.
Hello, my name is Sunghyun Park, but I go by Henry in English. I am an undergraduate student majoring in Biochemistry and Computer Science at Yonsei University in Seoul, South Korea.
I’ve been drawn to SYSTEMS since high school—first to the intricate systems of living cells. Over time that curiosity expanded beyond biology as I discovered how data-driven modeling can reveal structure in almost any complex process. The moment when AI-driven advances like AlphaFold reshaped structural biology and even sparked conversations about Nobel-level recognition crystallized something for me: AI isn’t just a tool for analysis. it can be an instrument of discovery. Today, I want to build models that blend mechanistic insight with machine learning, and I want to study how LLMs can reason faithfully and safely to support scientific practice.
Education.
-
 Mar. 2020 - Present
Mar. 2020 - PresentYonsei University
Biochemistry & Computer Science (Double Major)
GPA: 3.91/4.3 (CS Major GPA: 4.0/4.3)Military Service: Oct. 2022 - Jul. 2024 (R.O.K Air Force Sergeant)
Projects.
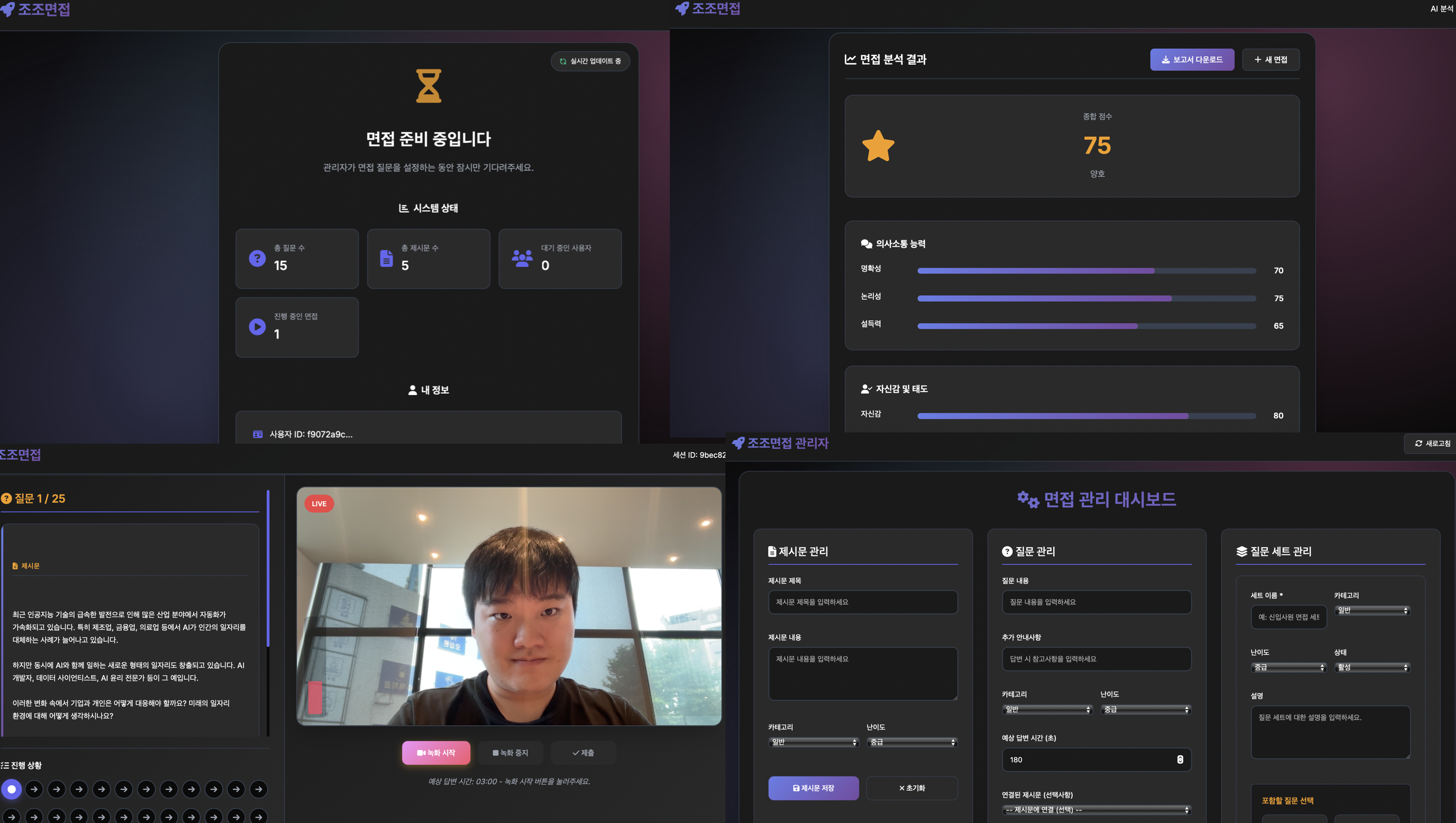
HACK SNU 2025 : JoJo Interview
We are developing the System architecture and AI engine for JoJo Interview, a service that enables high school students to practice interviews anytime, anywhere. We implement microservice architecture and build asynchronous pipelines for scalable video analysis. In terms of AI, we will design AI modules such as VLM/LLM for automated feedback and dynamic interview question generation.
Member: Jaeho Lim, Hwanhoon Chung, Sunghyun Park
My Academic Homepage
This Project is the website you are currently watching. The site includes features such as Gallery and Blog, in particular, the Blog supports search and parses Markdown files, allowing category-based search in a tree structure.
Member: Sunghyun Park
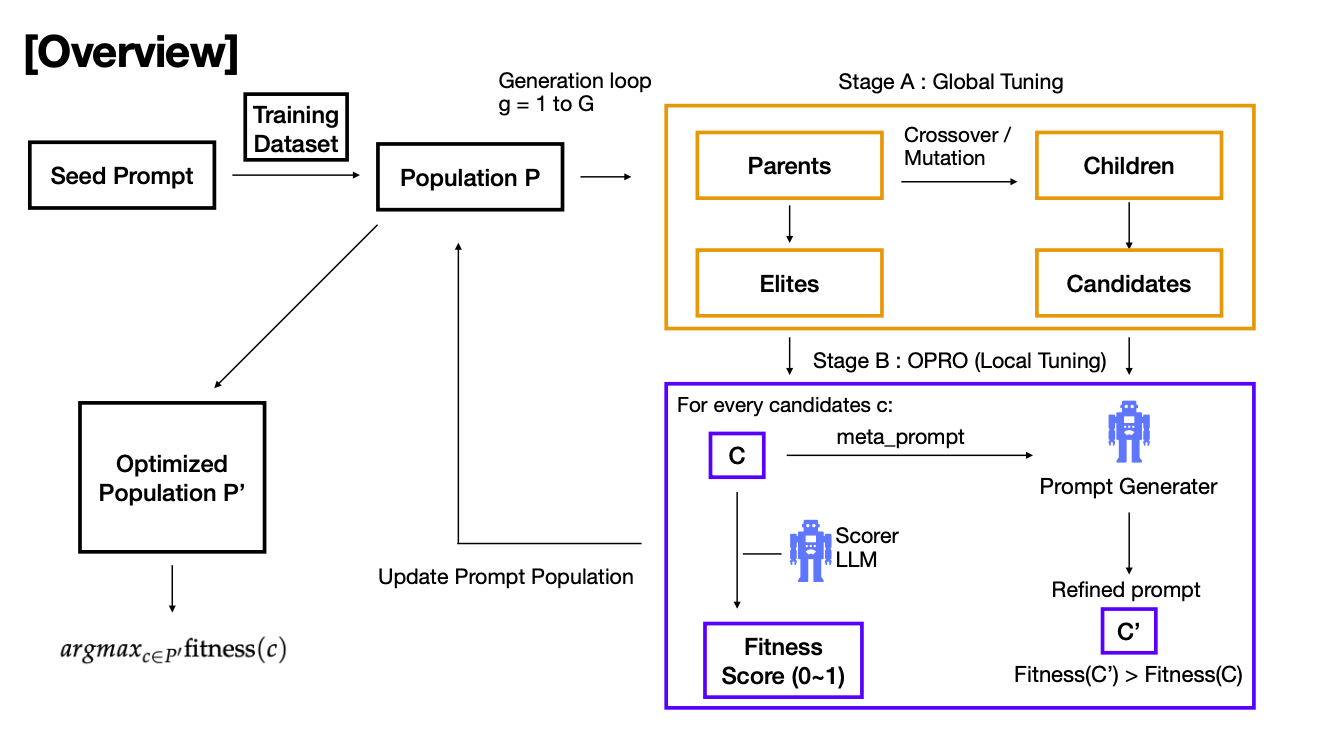
LLM Course Project : Prompt Optimization
We Implemented a prompt-optimization framework that combines OPRO with an evolutionary algorithm (selection, crossover, mutation) and an LLM-based scorer to drive self‑rewriting and selection and we evaluated on GSM8K with Llama-3.1-8B-Instruct and Mistral-7B-Instruct.
Member: Sunghyun Park
Reducing WebOS Boot Latency
Using ftrace and kernel instrumentation, we captured boot‑time function traces to identify hotspots and page faults. By reorganizing code sections via a custom linker script and optimizing the ELF binary layout, we reduced kernel page faults and improved boot times for embedded devices.
Members: Sunghyun Park, Jonghyun Hwang, Sungyoon Lim, Changwon Kim
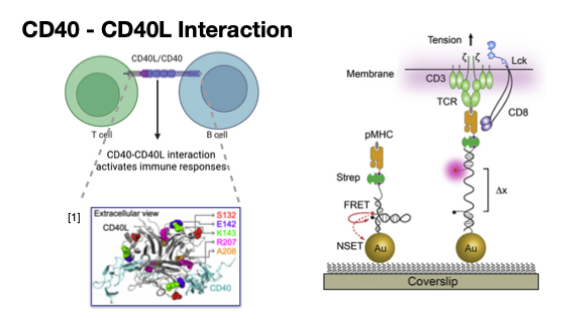
Force‑Driven Mechano‑Typing of B Cells - Multi-channel image analysis of CD40-CD40L interaction using autoencoder
We combined multi‑threshold molecular tension probes and three‑channel microscopy (DIC, RICM, TIRF) with a deep autoencoder to encode single‑cell images into 32‑D biophysical representations. This framework reveals how mechanical forces and CD40L mutations jointly shape B‑cell morphology and adhesion.
Members: Sunghyun Park, Yelim Jeon, Hyunkyu Choi
Topics of Interest.
Deep Learning Theory
I am deeply interested in the theoretical analysis of optimization and its dynamics in diffusion models. I’m currently studying the math behind these systems—clean SDE/ODE formulations and what they imply for convergence, stability, and generalization.
Large Language Models
I am interested in efficient and reliable inference (ex. speculative decoding, self-refinement), and alignment for faithfulness and safety (SFT/LoRA, DPO/ORPO). I also want to explore machine unlearning for LLMs—post-hoc removal of specific data or behaviors, auditing/attribution to trace sources, and principled “forgetting” guarantees to support safety, privacy, and dataset governance.
AI for Science
I want to build hybrid world models that blend data-driven learning with mechanistic domain models to produce accurate, robust, and sample-efficient simulators. Topics include physics-informed priors, system identification, data assimilation, and uncertainty calibration for scientific workflows.
Skills & Expertise.
Python & ML Frameworks
70%Python, PyTorch, TensorFlow
Deep Learning
65%Neural Networks, Transformers, Diffusion, LLM Alignment, LLM Fine-tuning
Computational Biology
70%Protein Struture Modeling, Microscopy Image Analysis
Data Analysis
60%Dimension Reduction, Unsupervised Clustering, Statistical Analysis
Experience.

In PoolC, I actively participated in AI-related paper studying groups and organized seminars on the mathematical foundations of deep learning. My seminars covered essential topics including information theory, maximum likelihood estimation (MLE), maximum a posteriori (MAP), and convex optimization, providing comprehensive understanding of the theoretical underpinnings of modern AI systems.

I serve as an academic mentor, guiding middle and high school students in mathematics learning and comprehensive academic planning. My role involves providing detailed guidance on major selection, offering comprehensive overviews of CS/AI and science major curricula, explaining prerequisites, and sharing effective study habits to help students make informed decisions about their academic futures.

My main research topic in Mechonbiology Lab was the investigation of Deep-Learning approach to make connection with AI and Biology, which involves multi-cell image analysis using compuatational methods.
Awards & Honors.

Honors Award, Yonsei - Spring 2025
Awarded to the top 10% students in Yonsei Students

Honors Award, Yonsei - Spring 2021
Awarded to the top 10% students in Yonsei Students
Contact.
- psh040@yonsei.ac.kr
- +82-10-9115-0823